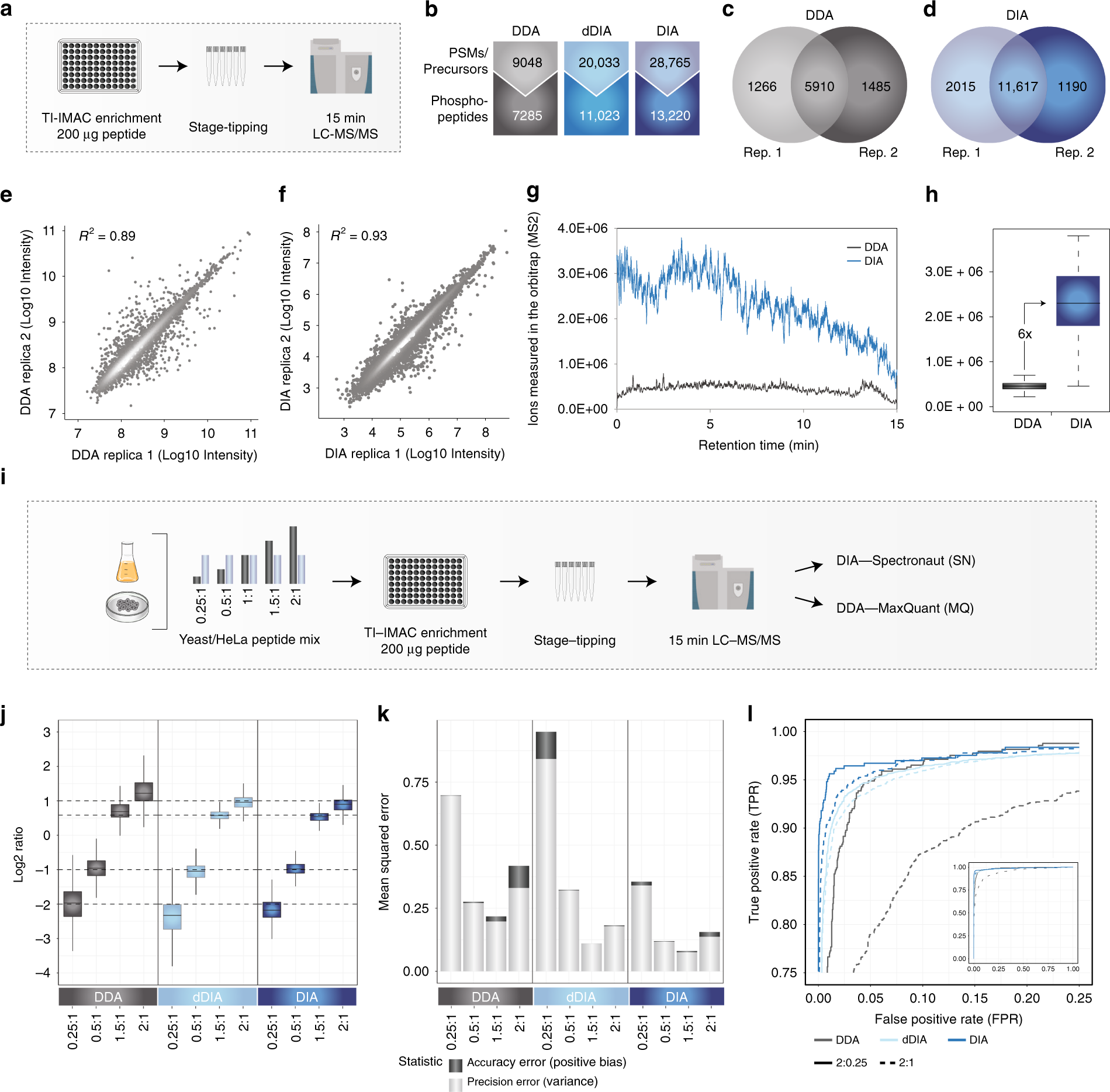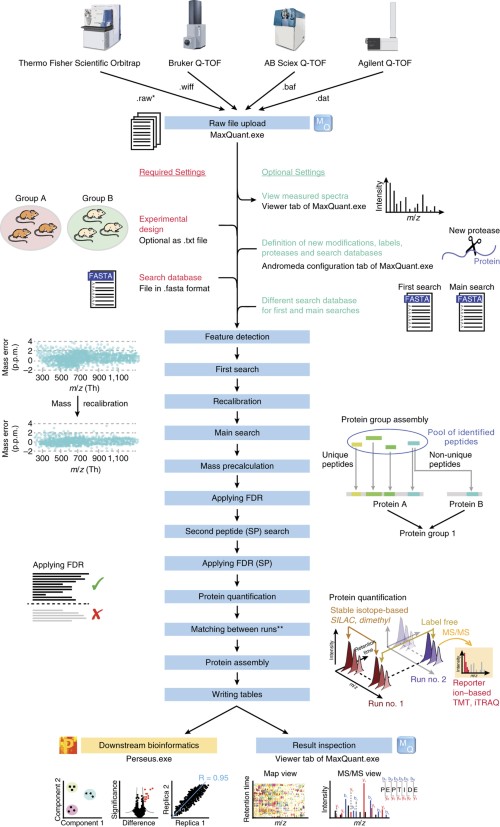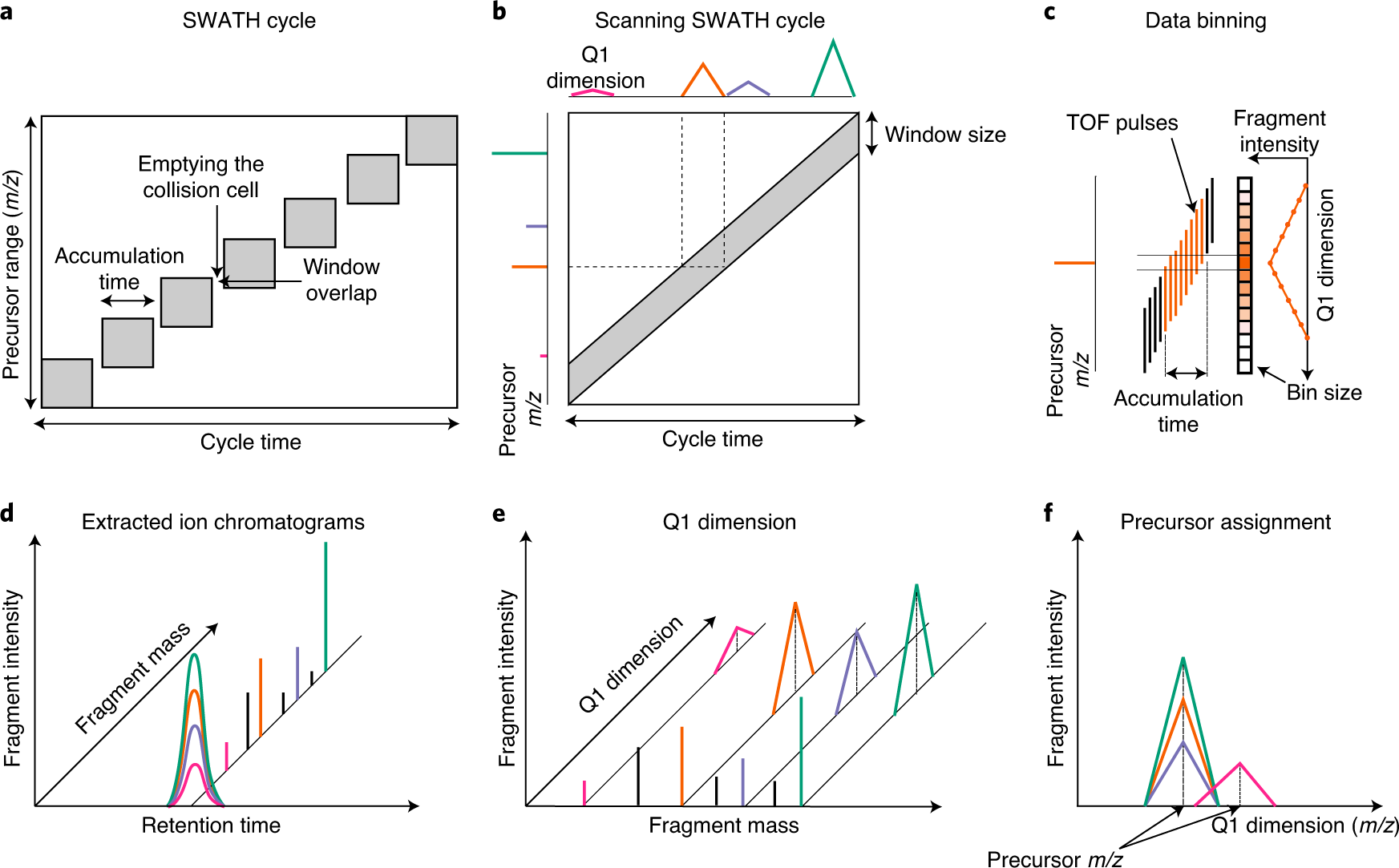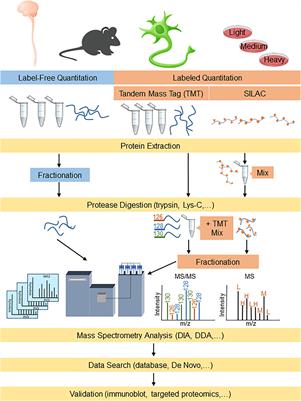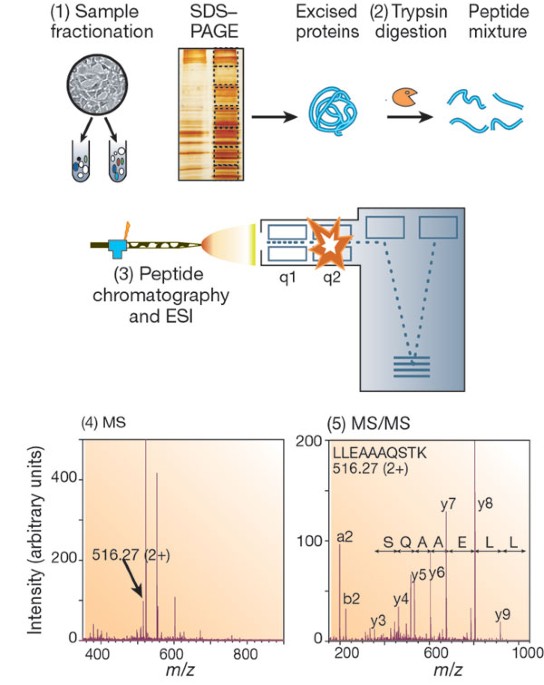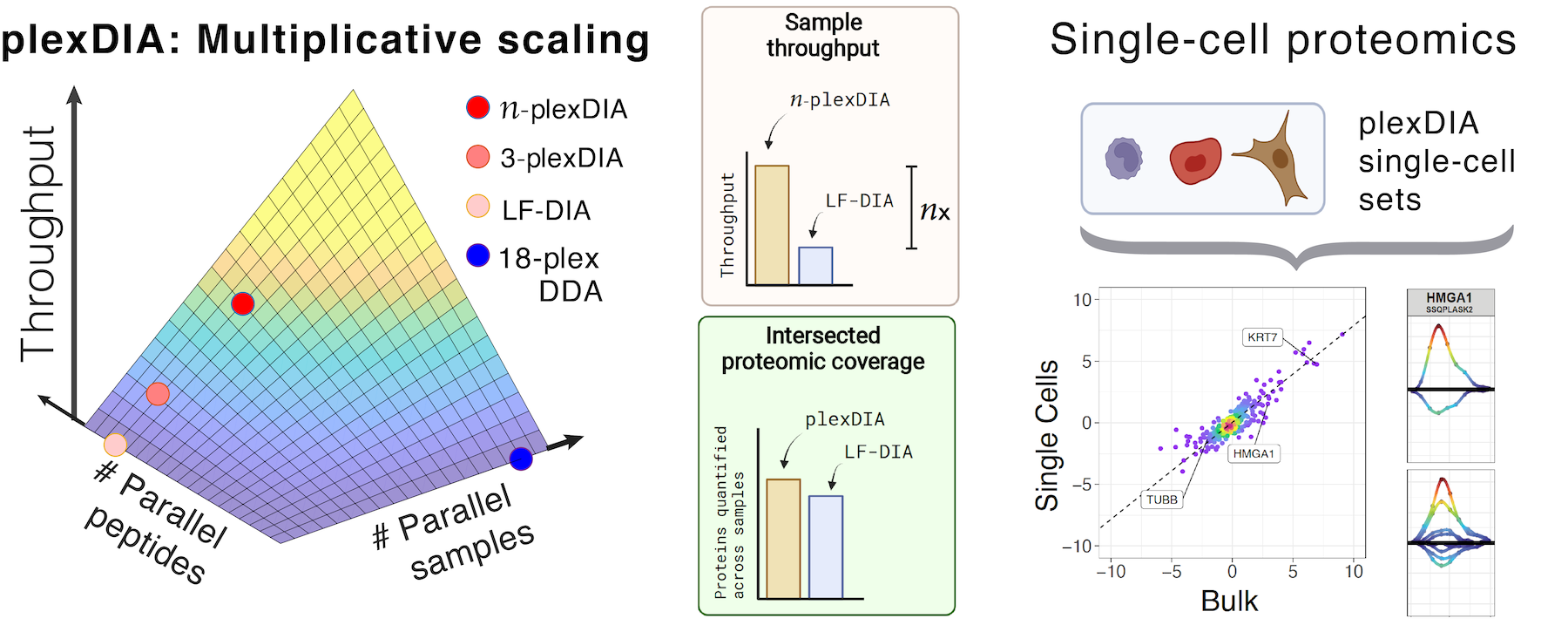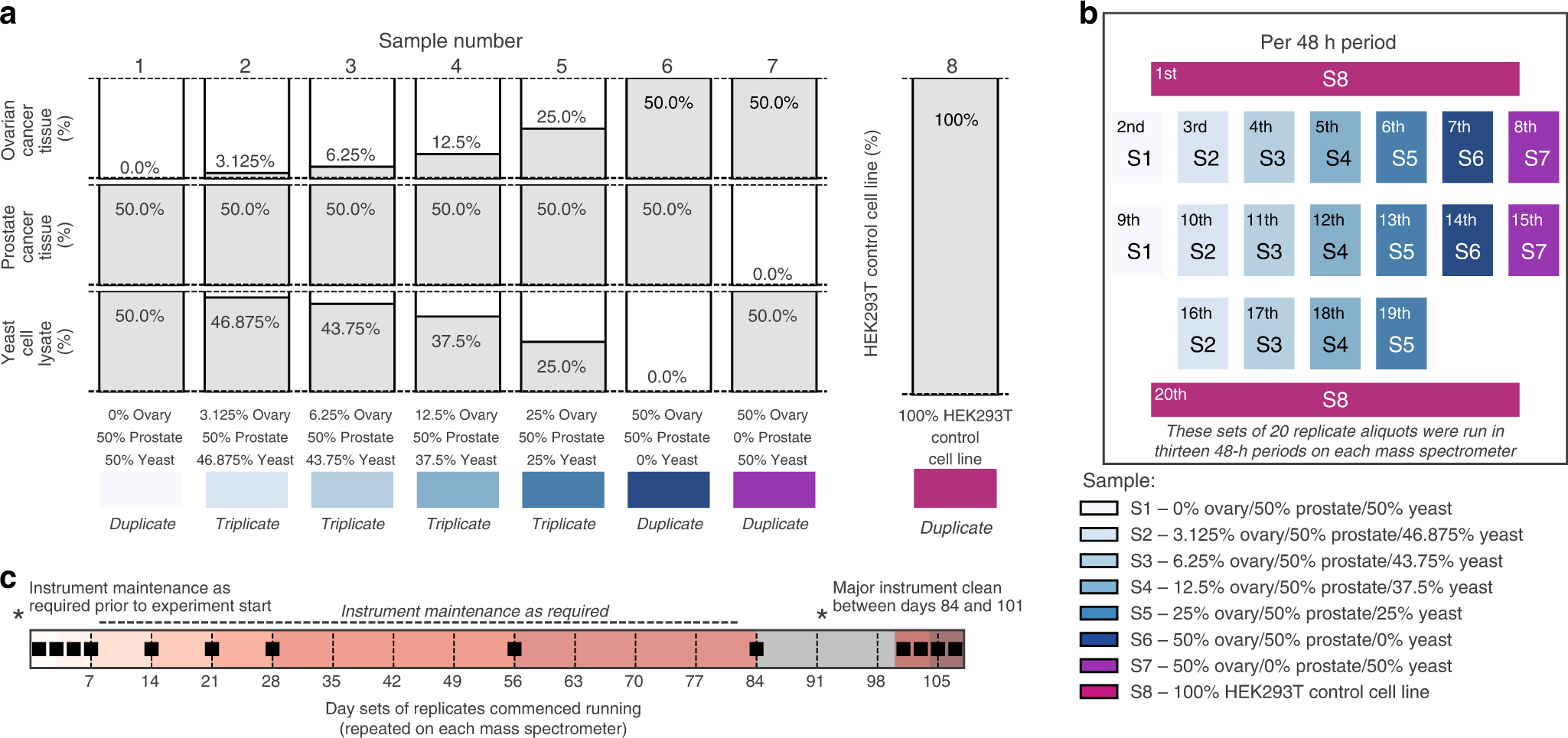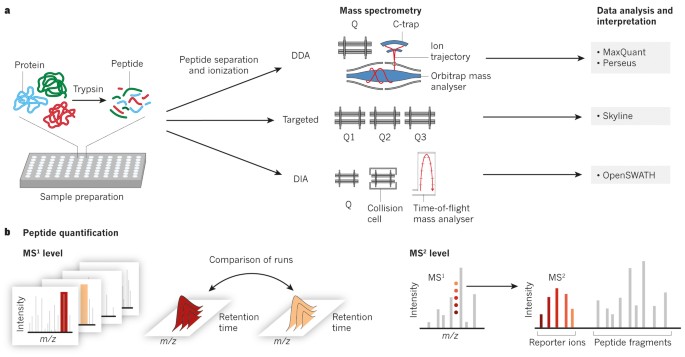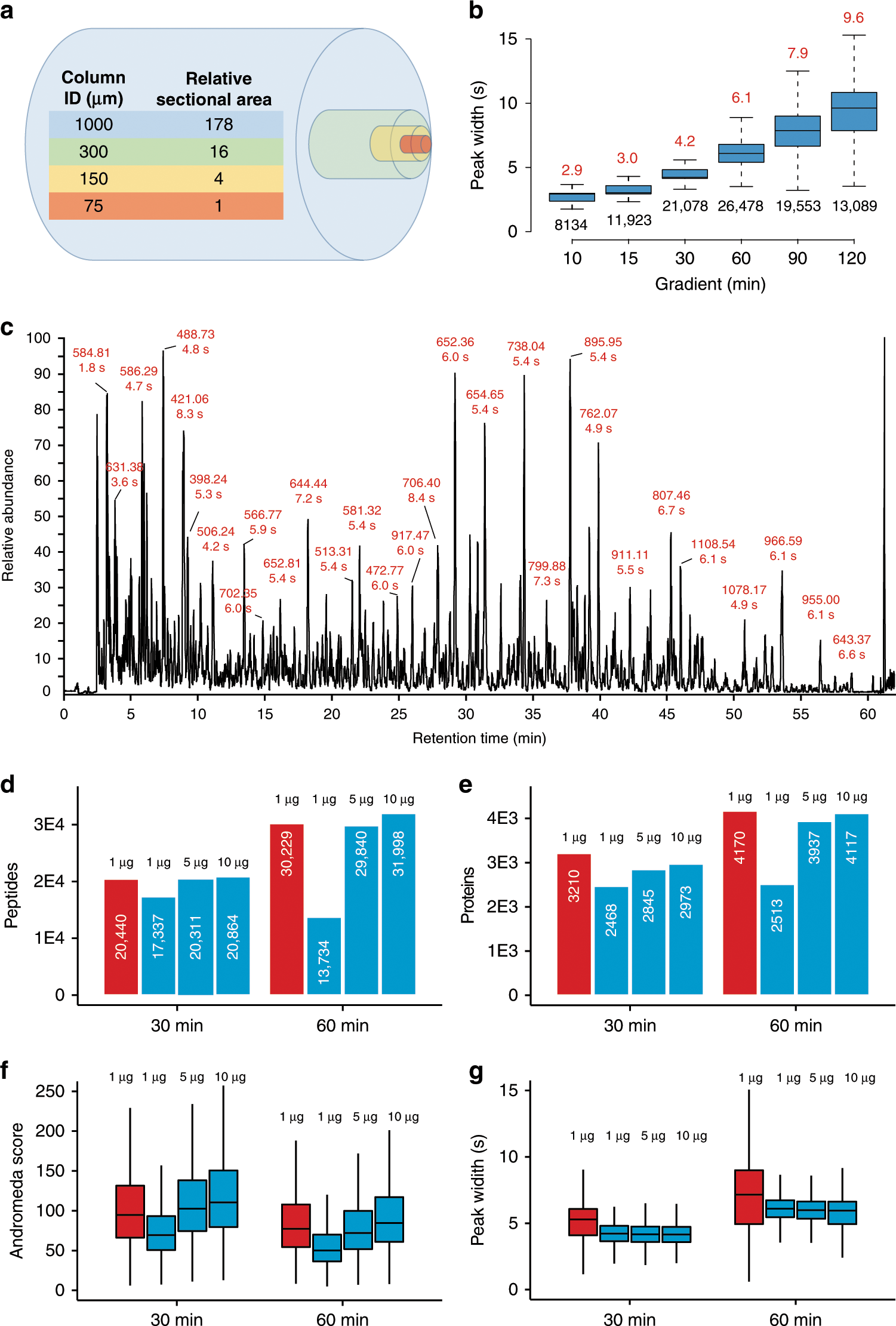
Robust, reproducible and quantitative analysis of thousands of proteomes by micro-flow LC–MS/MS | Nature Communications
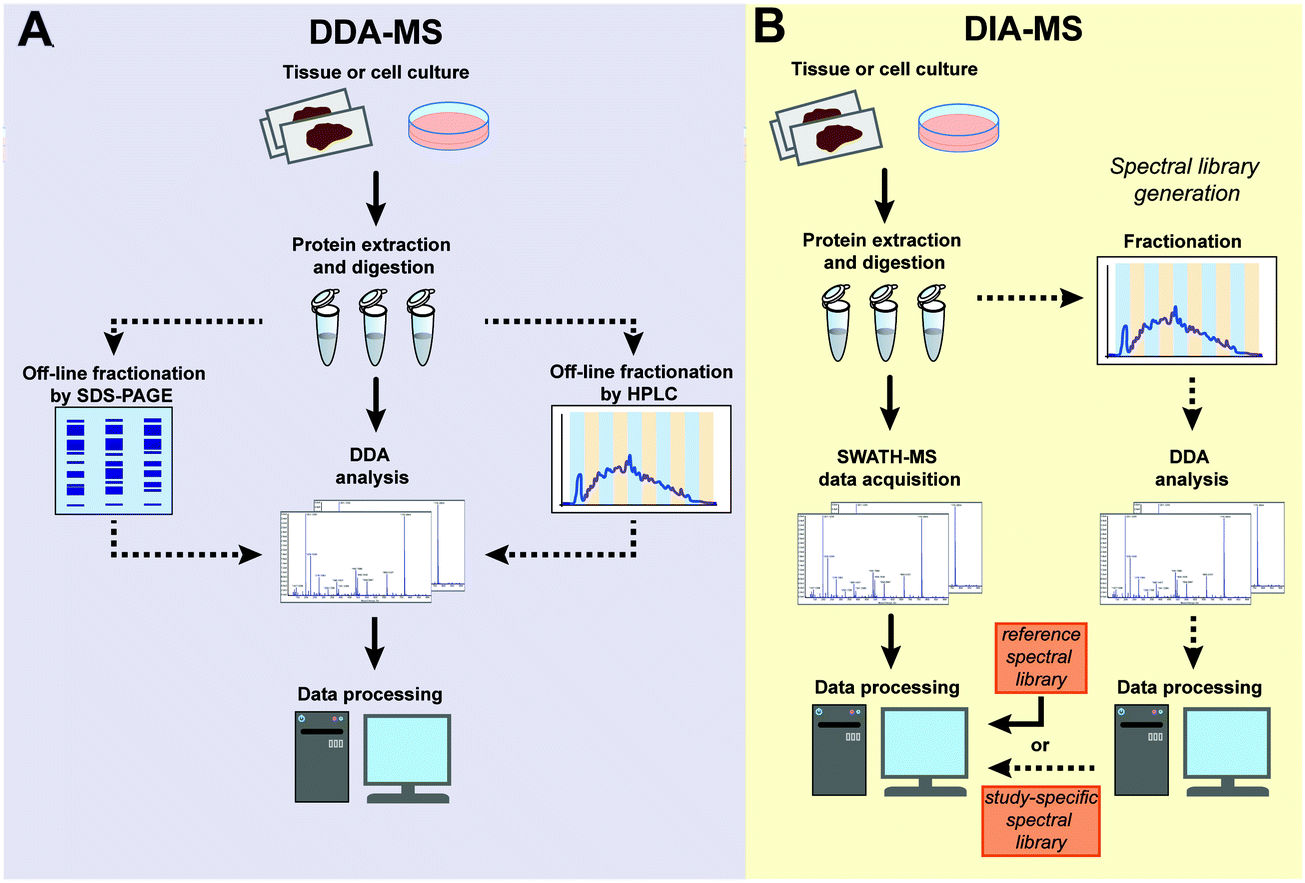
Data-independent acquisition mass spectrometry (DIA-MS) for proteomic applications in oncology - Molecular Omics (RSC Publishing) DOI:10.1039/D0MO00072H
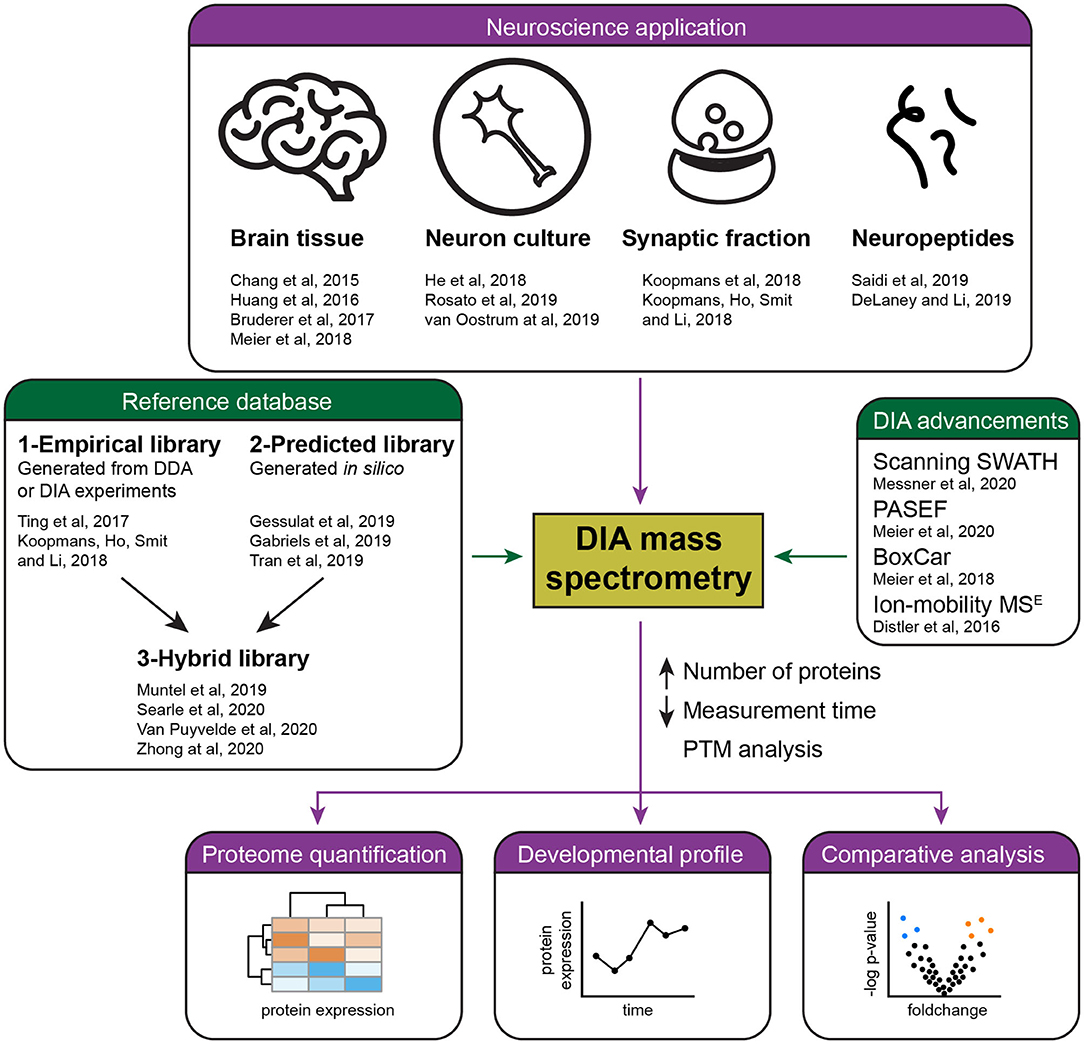
Frontiers | Recent Developments in Data Independent Acquisition (DIA) Mass Spectrometry: Application of Quantitative Analysis of the Brain Proteome

Hybrid Spectral Library Combining DIA-MS Data and a Targeted Virtual Library Substantially Deepens the Proteome Coverage - ScienceDirect
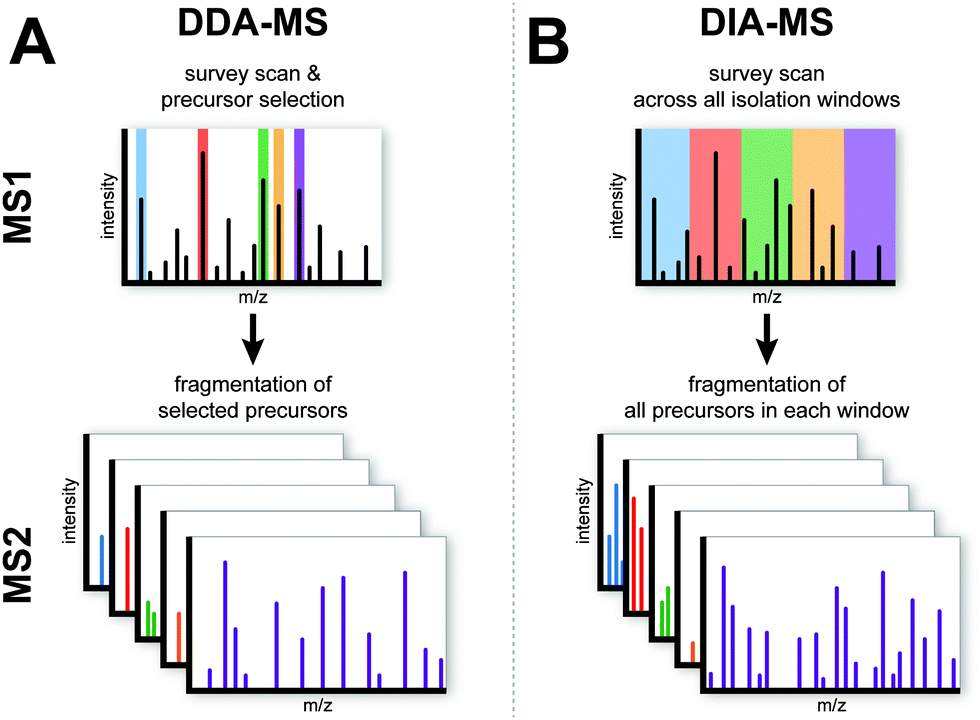
Data-independent acquisition mass spectrometry (DIA-MS) for proteomic applications in oncology - Molecular Omics (RSC Publishing) DOI:10.1039/D0MO00072H

DIA-NN: neural networks and interference correction enable deep proteome coverage in high throughput | Nature Methods
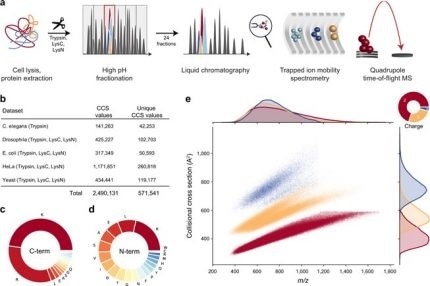
New Nature Communications publication by Mann & Theis Groups harnesses the benefits of large-scale peptide collisional cross section (CCS) measurements and deep learning for 4D-proteomics
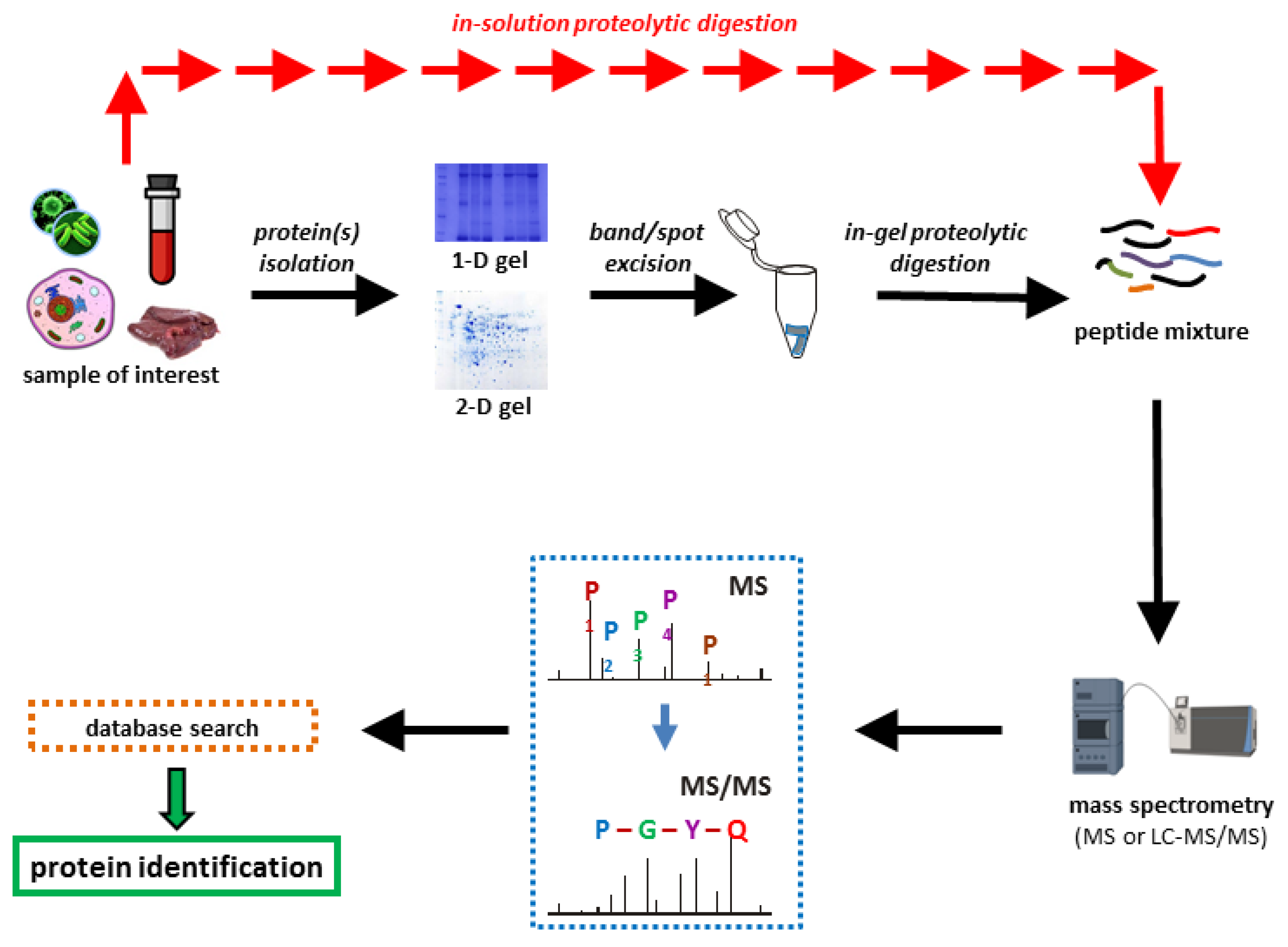
Proteomes | Free Full-Text | A Critical Review of Bottom-Up Proteomics: The Good, the Bad, and the Future of This Field
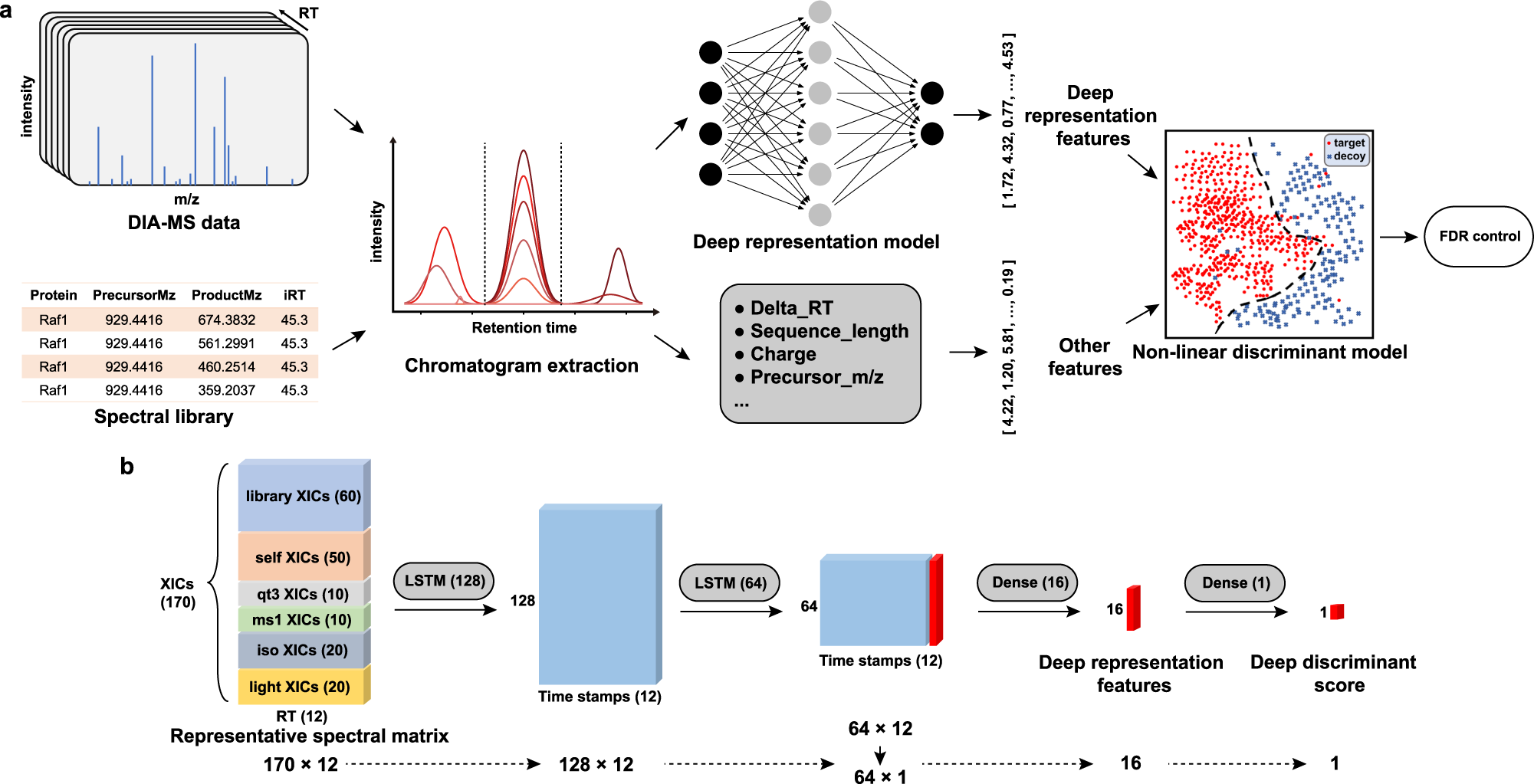
Deep representation features from DreamDIAXMBD improve the analysis of data-independent acquisition proteomics | Communications Biology
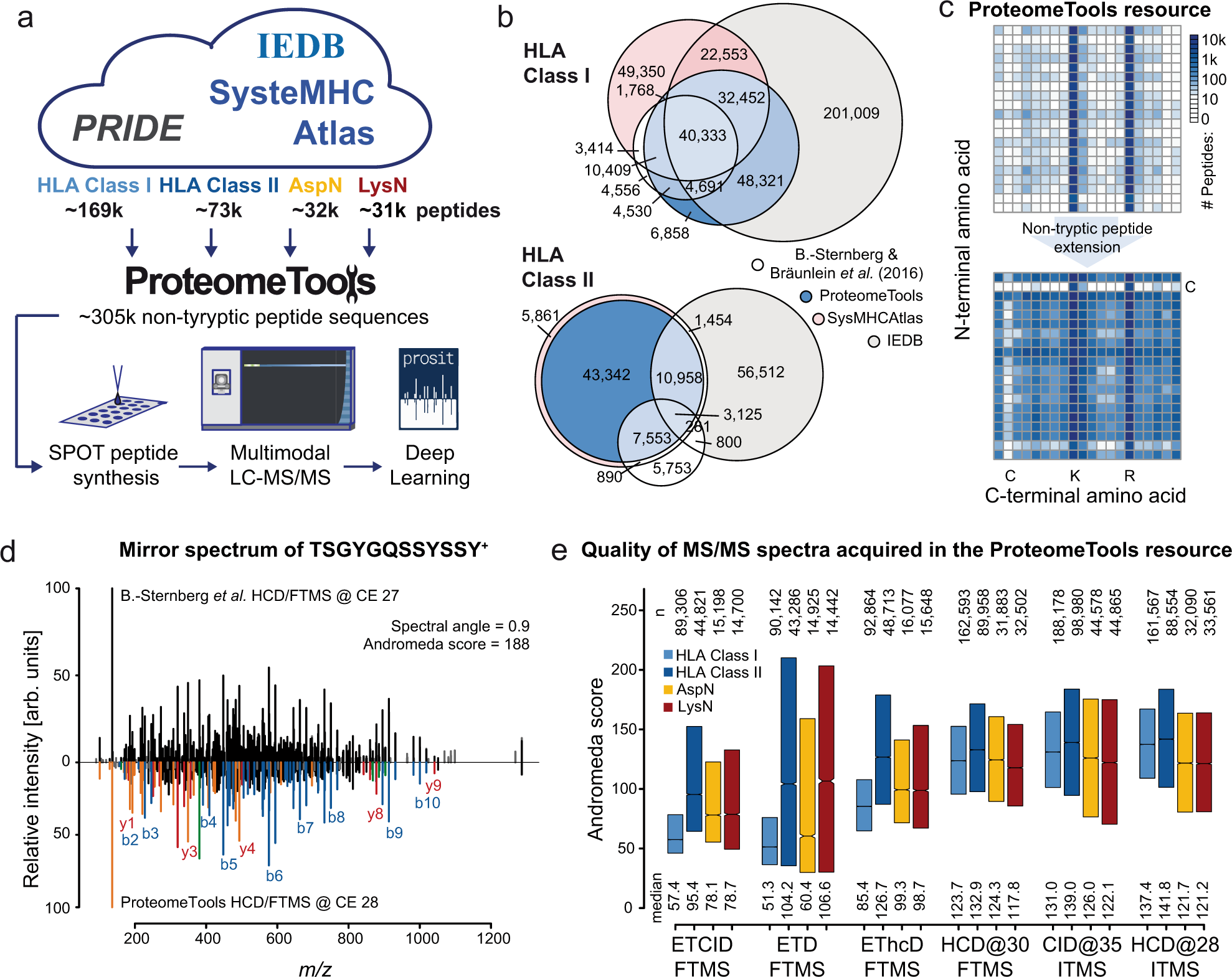
Deep learning boosts sensitivity of mass spectrometry-based immunopeptidomics | Nature Communications
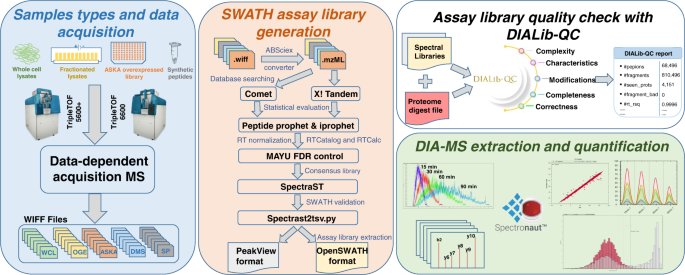
A comprehensive spectral assay library to quantify the Escherichia coli proteome by DIA/SWATH-MS | Scientific Data
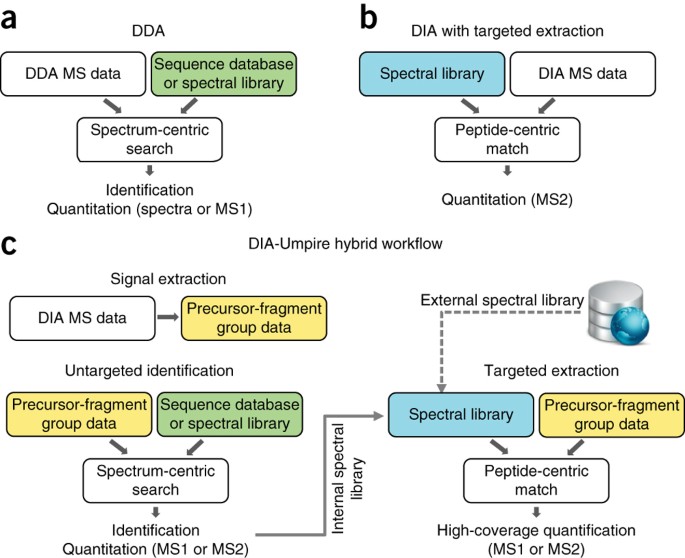
DIA-Umpire: comprehensive computational framework for data-independent acquisition proteomics | Nature Methods

DIAproteomics: A Multifunctional Data Analysis Pipeline for Data-Independent Acquisition Proteomics and Peptidomics | Journal of Proteome Research
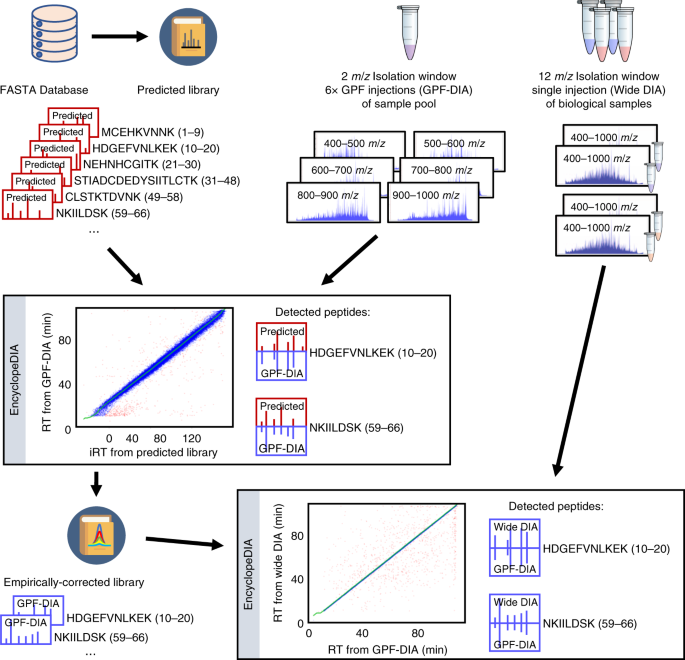
Generating high quality libraries for DIA MS with empirically corrected peptide predictions | Nature Communications

Deep Proteomics Using Two Dimensional Data Independent Acquisition Mass Spectrometry | Analytical Chemistry
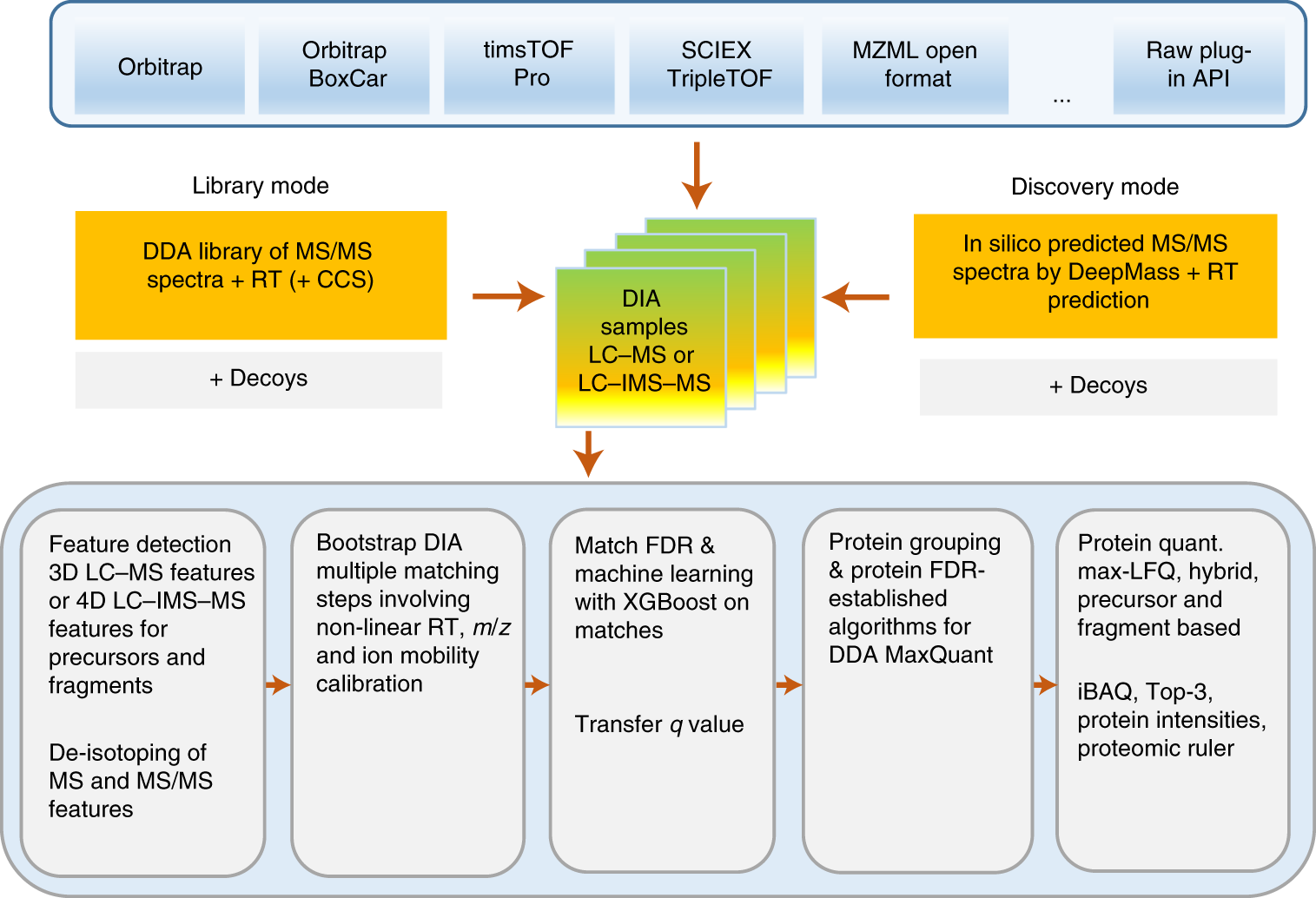
MaxDIA enables library-based and library-free data-independent acquisition proteomics | Nature Biotechnology

Chromatogram libraries improve peptide detection and quantification by data independent acquisition mass spectrometry | Nature Communications
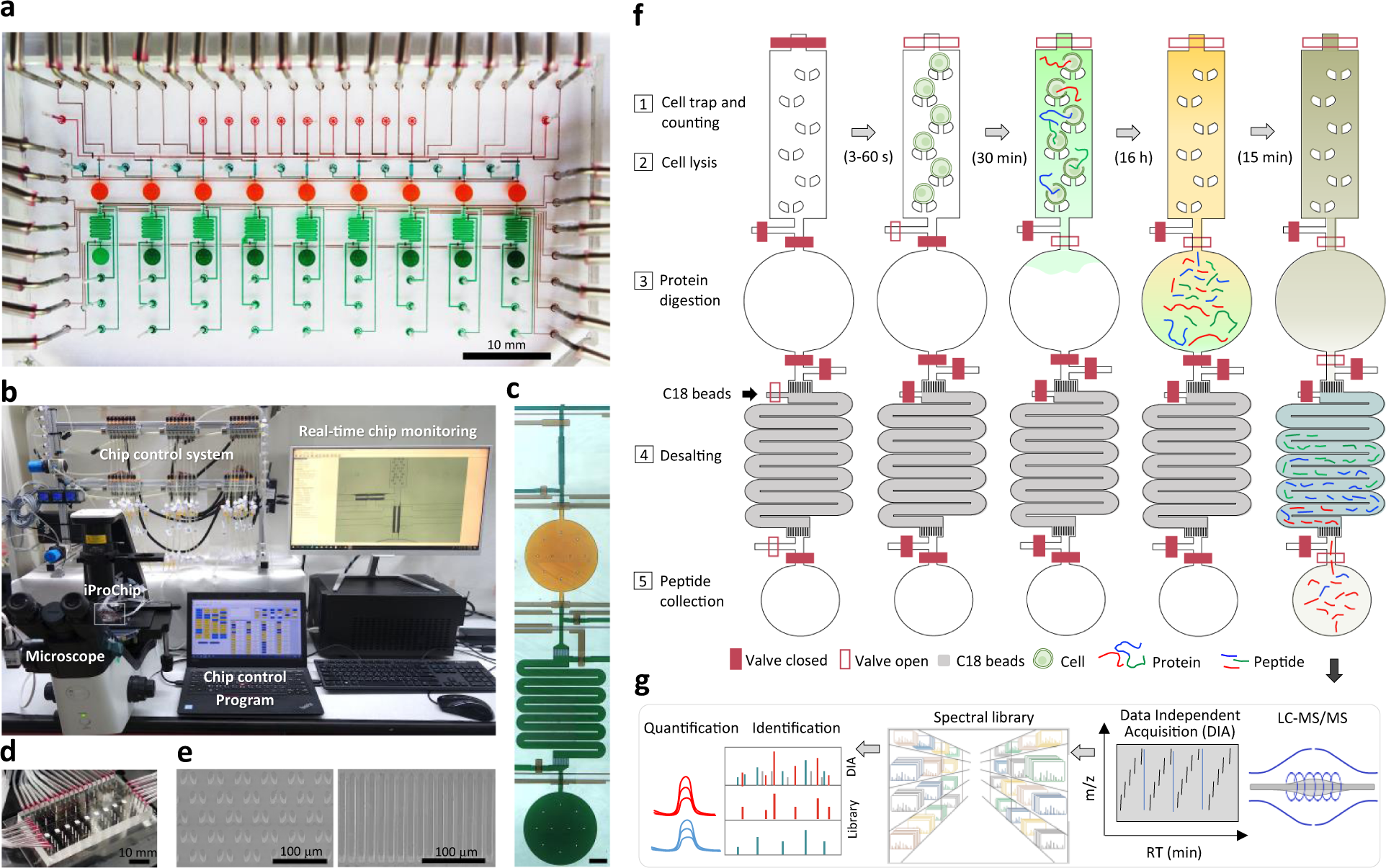
Streamlined single-cell proteomics by an integrated microfluidic chip and data-independent acquisition mass spectrometry | Nature Communications